Employing Nucleic Acid Conjugated Microspheres in the Field of Flow Cytometry: Part I
Posted: August 23, 2019
Analysis of nucleic acids using flow cytometry has become a more frequent practice. Horejsh et al. developed a fluid array system using microsphere-conjugated molecular beacons and flow cytometry for the specific, multiplexed detection of unlabeled nucleic acids in solution. To facilitate the system, molecular beacons were conjugated to microspheres using a biotin-streptavidin linkage. Spiro et al. describes a multiplexed, bead-based method that utilizes nucleic acid hybridizations on the surface of polystyrene microspheres to identify specific sequences in heterogeneous mixtures of DNA sequences, while Tang et al. employed imaging flow cytometry to analyze the self-assembly of DNA-conjugated polystyrene microspheres. This technique enables quantitative analysis of the assembly process and thereby enables detailed assessment of the effect of structural and process variables on the assembly yield. These studies illustrate the value of flow cytometry in nucleic acid research; but, how does one generate the bead nucleic acid complex described in these studies? We will be exploring the nuances of this process over the next few Flow Blogs. Here, we discuss the basic strategies employed. Please see the rest of our website for more detailed protocols.
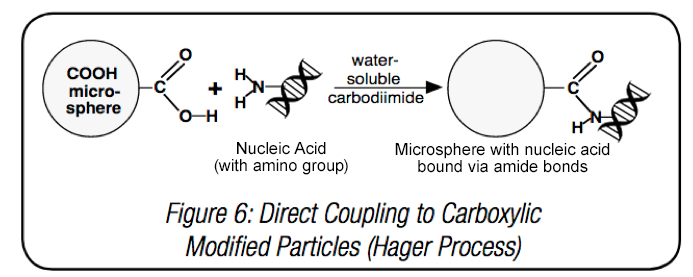
Several types of microspheres are utilized to isolate and purify nucleic acids. Both carboxylate-modified and streptavidin-coated microspheres, together with conjugated probes, can capture target sequences. Binding nucleic acids to microparticles directly using carboxyl surface groups features a coupling process similar to that of proteins. One of the most effective coupling chemistries uses -COOH groups on carboxylate-modified latex (CML) microspheres to conjugate amine modified DNA to the bead surface, via water soluble carbodiimide (WSC) chemistry, or EDC.
Another common approach is to use a binding strategy that features the Streptavidin-Biotin complex. Biotinylated oligonucleotides are a useful tool in many fields of biological research, particularly as probes for hybridization studies. Both strategies feature single-point attachment of the probe (which may or may not including a spacing sequence or chain) to permit hybridization of the sequence. Stay tuned for additional tips as we will further discuss the optimization of conjugation protocols and effects of varying bead types and binding strategies in upcoming blogs.
Horejsh, D. (2005). A molecular beacon, bead-based assay for the detection of nucleic acids by flow cytometry. Nucleic Acids Research, 33(2). doi:10.1093/nar/gni015
Spiro, A., Lowe, M., & Brown, D. (2000). A Bead-Based Method for Multiplexed Identification and Quantitation of DNA Sequences Using Flow Cytometry. Applied and Environmental Microbiology, 66(10), 4258-4265. doi:10.1128/aem.66.10.4258-4265.2000
Tang, H., Deschner, R., Allen, P., Cho, Y., Sermas, P., Maurer, A., . . . Willson, C. G. (2012). Analysis of DNA-Guided Self-Assembly of Microspheres Using Imaging Flow Cytometry. Journal of the American Chemical Society, 134(37), 15245-15248. doi:10.1021/ja3066896
