Employing Nucleic Acid Conjugated Microspheres in the Field of Flow Cytometry: Part II
Posted: September 3, 2019
Last week we discussed how nucleic acid attachment to microspheres for subsequent flow cytometry analysis has become an important technique in biotechnology research and development. Beads and attached ligands are utilized by both researchers and clinicians alike, and are employed in such methods as DNA sequencing, as aptamers in protocols, to Triple-Helix-Mediated-Affinity Capture, and others. In our previous blog we discussed different approaches for attaching your nucleic acid. This week we want to focus on optimizing the concentration of DNA for better performance.
Using flow cytometry, Wittebolle et al. performed a series of assays exploring the input of varied concentrations of DNA when coupling to 3µm microspheres. The optimal concentration was determined to be 15pmol of oligo for every 1x1015 beads. This resulted in an oligo surface density of 336 oligo/µm2. Concentrations above and below this value showed less conjugation efficiency and is attributed to steric effects. Steric effects can sometimes be diminished by including a spacer onto the bead surface, but often times this does not significantly improve the assay.
So, how does one calculate the density?
- Determine the number of beads/mg to be used in the experiment.
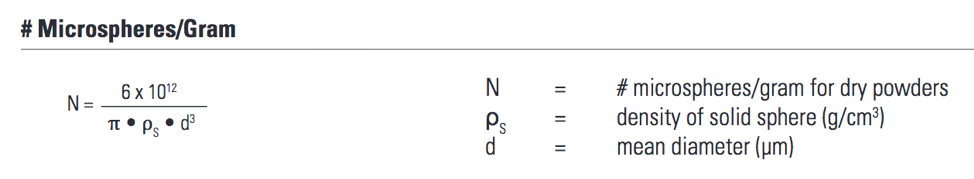
- Determine the mass you want in the experiment.
- Find the area for one sphere.

- Multiply the surface area for one sphere by the number of spheres to be used in the experiment. This is your total surface area.
- Determine the mass of oligo to add in order to achieve ~336 oligo/bead.
Taking these few extra steps and performing these calculations in advance could significantly improve your flow results when analyzing bead-oligo conjugations and save on reagent cost during protocol optimization.
Wittebolle, L., Verstuyft, K., Verstraete, W., & Boon, N. (2006). Optimisation of the amino–carboxy coupling of oligonucleotides to beads used in liquid arrays. Journal of Chemical Technology & Biotechnology, 81(3), 476–480. doi: 10.1002/jctb.1421
